In jMRUI user can find algorithms that bear with simulation of spin systems :
NMR-SCOPE
NMR-SCOPE is a well-established plugin for basic simulation of metabolite signals. It supports batch processing of multiple metabolite spectra and possesses a familiar graphical interface.
References
- Stefan, D., Di Cesare, F., Andrasescu, A., Popa, E., Lazariev, A., Vescovo, E., Strbak, O., Williams, S., Starcuk, Z., Cabanas, M., van Ormondt, D., Graveron-Demilly. D. Quantitation of magnetic resonance spectroscopy signals: the jMRUI software package. Measurement Science and Technology 20:104035 (9 pp), 2009. doi: 10.1088/0957-0233/20/10/104035
NMRScopeB
NMRScopeB (current version 2.0, distributed together with jMRUI) is a versatile simulator of multiple metabolite spectra. The plugin provides the functionality useful for the simulation of coupled spin systems during the NMR experiment. In the simulation, properties such as chemical shifts, spin-spin coupling, relaxation, spatial and/or spectral excitation selectivity, and customized pulse sequences are accounted for. The primary target is to support the simulation of metabolite FID signals in biomedical MR spectroscopy, as needed for spectroscopic quantitation, but many functions are meant to support the development of methods for MR spectroscopy or spectroscopic imaging, or education.
New in version 2.0: – platform independence improved by reimplementation of the kernel in Python (connected to jMRUI via sockets), – the simulator is available under both Linux and Windows, – the new user interface facilitates the selection and simulation of all metabolites selected for the basis in a single step, – transparent display of pulse sequences, – graphs of the simulated signals can be stored in vector graphics formats, e.g. metafiles, svg etc. for lossless exportation to documents,- the simulation can be launched by macros , – a new protocol for the SPECIAL sequence, – the simulated FIDs can be integrated and/or multiplied by a user defined function (for simulation of VOI selection, inhomogeneous excitation, chemical-shift effect), – the simulated metabolites can be saved in a metabolite list and loaded directly in QUEST or AQSES as a new basis set (no need to create a list of metabolites manually from individually simulated metabolites).
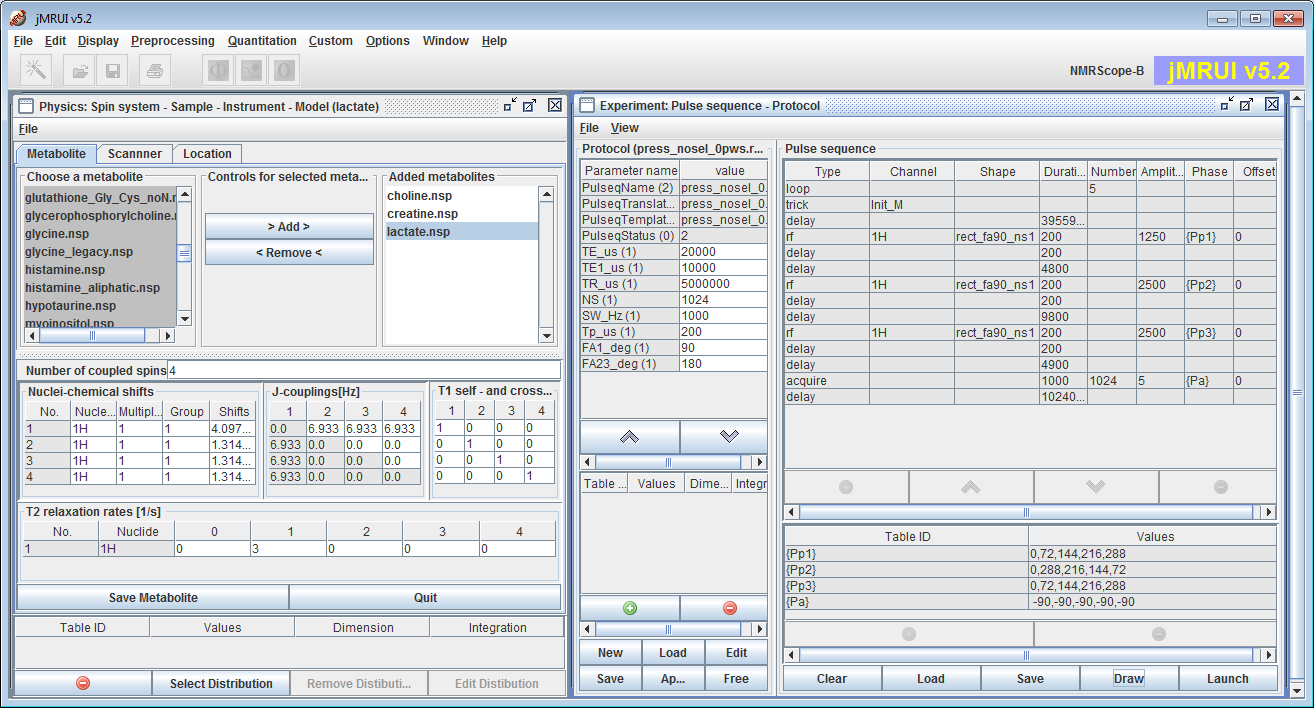
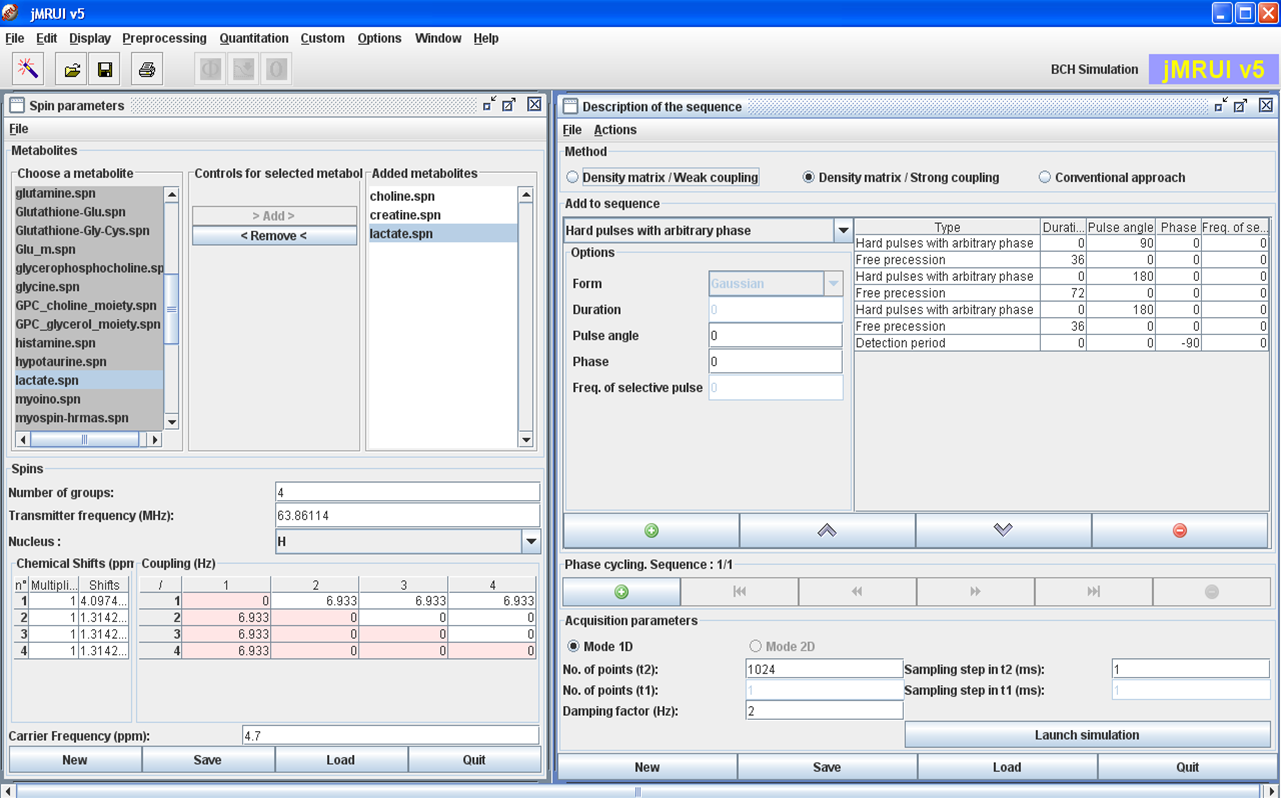
Typical interface with two main panels: the Physics and the Experiment panels.
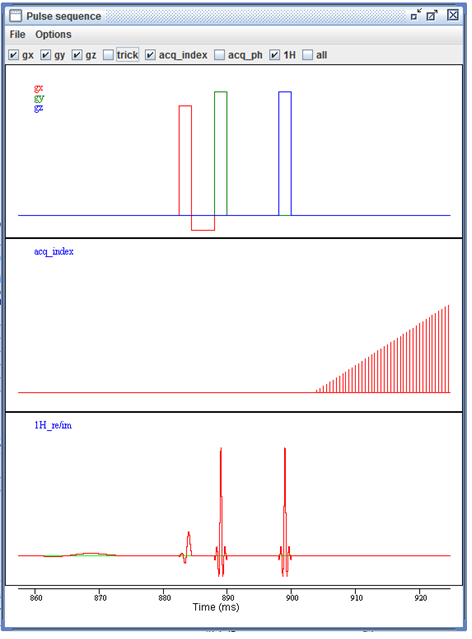
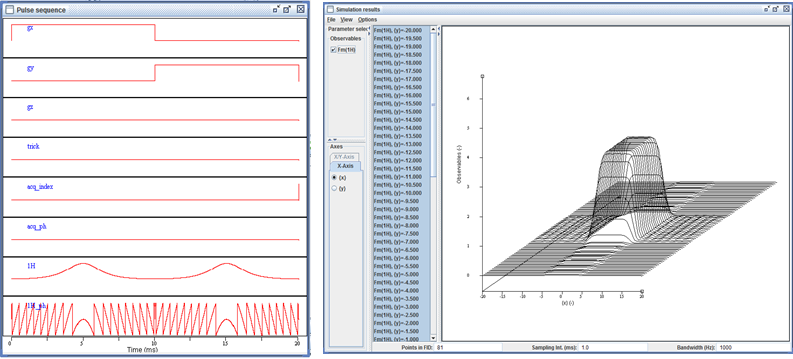
The interactive pulse sequence display.
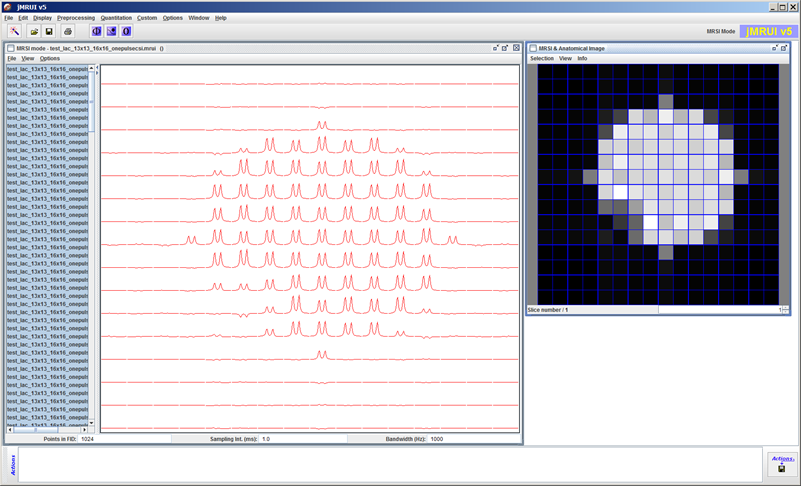
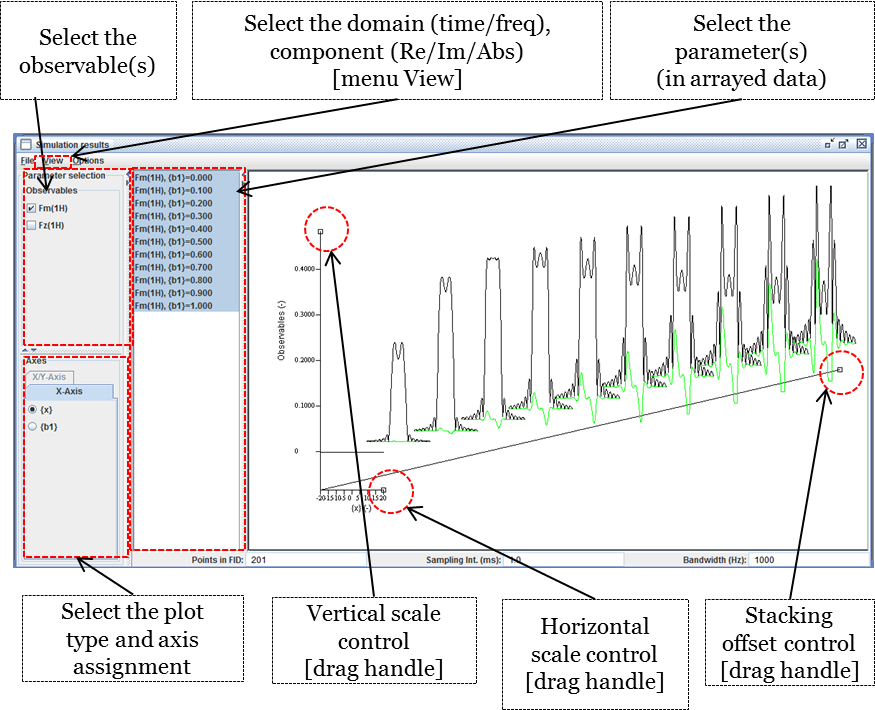
The extended signal/spectrum browser lets the user inspect multiparametric simulation results, such as here the dependence of the excitation profile on flip angle.
References
- Z Starcuk Jr, J Starcukova; Quantum-mechanical simulations for in vivo MR spectroscopy: Principles and possibilities demonstrated with the program NMRScopeB. Anal Biochem. DOI: 10.1016/j.ab.2016.10.007.
- Z Starcuk Jr, J Starcukova, O Strbak and D Graveron-Demilly: Simulation of coupled-spin systems in the steady-state free-precession acquisition mode for fast magnetic resonance (MR) spectroscopic imaging. Meas. Sci. Technol. 20, 10, 104033 (2009). doi: 10.1088/0957-0233/20/10/104033
Spectrum synthesizer
Artificial spectra with arbitrary signals can be synthesised from a list of frequencies, amplitudes, dampings and phases describing the signals in the spectrum. Then, noise with a known average value and standard deviation can be added to the artificial spectra. These artificial spectra are useful to test and compare the performance of the algorithms, for instance, with increasing levels of noise or in the presence of artefacts.